UT’s Rising Stars of Microbiology

by Logan Judy

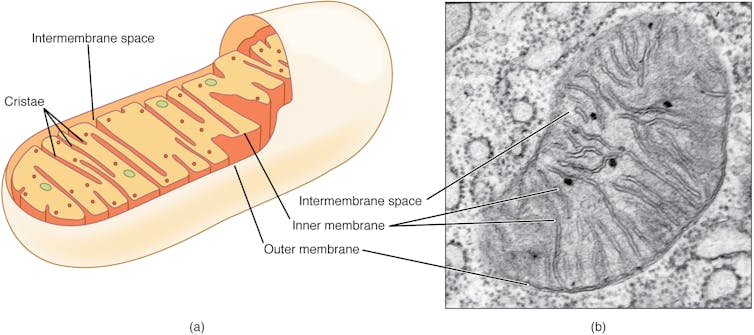
Mitochondria have primarily been known as the energy-producing components of cells. But scientists are increasingly discovering that these small organelles do much more than just power cells. They are also involved in immune functions such as controlling inflammation, regulating cell death and responding to infections.
Research from my colleagues and I revealed that mitochondria play another key role in your immune response: sensing bacterial activity and helping neutrophils, a type of white blood cell, trap and kill them.
For the past 16 years, my research has focused on understanding the decisions immune cells make during infection and how the breakdown of these decision-making processes cause disease. My lab’s recent findings shed light on why people with autoimmune diseases such as lupus may struggle to fight infections, revealing a potential link between dysfunctional mitochondria and weakened immune defenses.
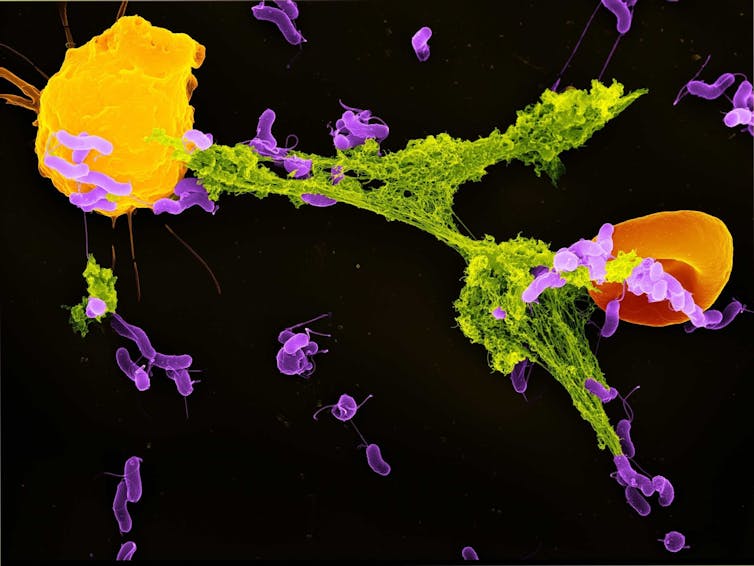
Neutrophils are the most abundant type of immune cell and serve as the immune system’s first responders. One of their key defense mechanisms is releasing neutrophil extracellular traps, or NETs – weblike structures composed of DNA and antimicrobial proteins. These sticky NETs trap and neutralize invading microbes, preventing their spread in the body.
Until recently, scientists believed that NET formation was primarily triggered by cellular stress and damage. However, our study found that mitochondria can detect a specific bacterial byproduct – lactate – and use that signal to initiate NET formation.
Lactate is commonly associated with muscle fatigue in people. But in the context of bacterial infections, it plays a different role. Many bacteria release lactate as part of their own energy production. My team found that once bacteria are engulfed by a compartment of the cell called the phagosome, neutrophils can sense the presence of this lactate.
Inside the phagosome, this lactate communicates to the neutrophil that bacteria are present and that the antibacterial processes are not sufficient to kill these pathogens. When the mitochondria in neutrophil cells detect this lactate, they start signaling for the cell to get rid of the NETs that have entrapped bacteria. Once the bacteria are released outside the cell, other immune cells can kill them.
When we blocked the mitochondria’s ability to sense lactate, neutrophils failed to produce NETs effectively. This meant bacteria were more likely to escape capture and proliferate, showing how crucial this mechanism is to immune defense. This process highlights an intricate dialogue between the bacteria’s metabolism and the host cell’s energy machinery.
What makes this finding surprising is that the mitochondria within cells are able to detect bacteria trapped in phagosomes, even though the microbes are enclosed in a separate space. Somehow, mitochondrial sensors can pick up cues from within these compartments – an impressive feat of cellular coordination.
Our study is part of a growing field called immunometabolism, which explores how metabolism and immune function are deeply intertwined. Rather than viewing cellular metabolism as strictly a means to generate energy, researchers are now recognizing it as a central driver of immune decisions.
Mitochondria sit at the heart of this interaction. Their ability to sense, respond to and even shape the metabolic environment of a cell gives them a critical role in determining how and when immune responses are deployed.
For example, our findings provide a key reason why patients with a chronic autoimmune disease called systemic lupus erythematosus often suffer from recurrent infections. Mitochondria in the neutrophils of lupus patients fail to sense bacterial lactate properly. As a result, NET production was significantly reduced. This mitochondrial dysfunction could explain why lupus patients are more vulnerable to bacterial infections – even though their immune systems are constantly activated due to the disease.
This observation points to mitochondria’s central role in balancing immune responses. It connects two seemingly unrelated issues: immune overactivity, as seen in lupus, and immune weakness like increased susceptibility to infection. When mitochondria work correctly, they help neutrophils mount an effective, targeted attack on bacteria. But when mitochondria are impaired, this system breaks down.
Our discovery that mitochondria can sense bacterial lactate to trigger NET formation opens up new possibilities for treating infections. For instance, drugs that enhance mitochondrial sensing could boost NET production in people with weakened immune systems. On the flip side, for conditions where NETs contribute to tissue damage – such as in severe COVID-19 or autoimmune diseases – it might be beneficial to limit this response.
Additionally, our study raises the question of whether other immune cells use similar mechanisms to sense microbial metabolites, and whether other bacterial byproducts might serve as immune signals. Understanding these pathways in more detail could lead to new treatments that modulate immune responses more precisely, reducing collateral damage while preserving antimicrobial defenses.
Mitochondria are not just the powerhouses of the cell – they are the immune system’s watchtowers, alert to even the faintest metabolic signals of bacterial invaders. As researchers’ understanding of their roles expands, so too does our appreciation for the complexity – and adaptability – of our cellular defenses.
Andrew Monteith, Assistant Professor of Microbiology, University of Tennessee
This article is republished from The Conversation under a Creative Commons license. Read the original article.
by Logan Judy

by Logan Judy

by Logan Judy

by Logan Judy

by Logan Judy

by Hanna Boshnag
Microplastics seems to be a big buzzword these days. It’s easy to roll one’s eyes at another sensational news headline, but these could have truly harmful impacts on the environment. The use of plastic mulching in agriculture could alter the soil microbiome and have adverse effects on the broader ecosystem. Biodegradable mulch is a potential alternative, but it’s uncertain how significant of a difference it makes.
Toyosi Nimat Ajide-Bamigboye, a PhD candidate at the University of Tennessee, Knoxville, is working to characterize this difference.
“Both biodegradable and non-biodegradable mulches generate fragments in the environment, which I study to see how it affects microorganism growth and function,” she said.
To ensure they are not inadvertently releasing microplastics into the environment, the lab collects soil from a field and uses a greenhouse as a controlled environment. After mulch is laid down, Ajide-Bamigboye checks a variety of factors over the course of six months: soil pH, degradation, and electrical conductivity. Once a month, a sample is sent off for sequencing to map changes in the microbiome.
This is a long and tedious process, which Ajide-Bamigboye says she enjoys every second of. She is passionate about every part of the process of doing research because it’s a working step to discovering something new.
Ajide-Bamigboye grew up in Nigeria, where she studied microbiology for both her bachelor’s and master’s degrees. When she decided to pursue her PhD at UT, she brought her husband and two children with her. Despite the difficulties of moving her family, she credits them as the reason she’s able to wholeheartedly work on her research.
“I have a five-year-old daughter and three-year old son,” she said. “My husband is my support system, and when I’m in school, I know I don’t have to worry about them because I know he has it under control.”
After completing her PhD, Ajide-Bamigboye hopes to stay with her family here and work in industry, using her research and skills to contribute creative solutions in the field of agricultural microbiology.
by Logan Judy


by Hanna Boshnag
Having been involved in conservation in high school, Liz Glasgo was excited to study biology as an undergraduate. When she got to Bowling Green State University, her catalyst for choosing microbiology was learning about marine microbes: diatoms, dinoflagellates, and radiolarians.
“I thought they were so interesting and beautiful to look at,” she said.
Later her undergraduate research focused on freshwater microbiology. After completing her bachelor’s degree, she chose to come to the University of Tennessee because of the sense of community within the Department of Microbiology.
Glasgo is a part of the Zinser Lab, where her research centers on Vibrio natriegens, a fast-growing, Gram-negative marine bacterium. She is attempting to characterize its oxidative stress response and fitness outcomes following genome reduction.
“V. natriegens is an emerging model organism, and the developments of new techniques and a better understanding of its physiology will be helpful to the scientific community,” she said.
For much of microbiology’s history, Escherichia coli has been the primary model organism for researchers to develop new techniques and understand a variety of biological systems. Discoveries as important as the genetic code, DNA replication, gene regulation, and more have been made possible with E. coli as the go-to organism for microbial study.
So why the potential switch to V. natriegens? The difference in growth rate. The marine bacterium has the fastest known generation time and is much quicker to double when compared with E. coli. This shortened growth time introduces the potential of faster experimentation and advanced genomic techniques. Glasgo’s work to better understand V. natriegens genetics and physiology is, thus, all the more important.
To Glasgo, the most exciting aspect of research is directly carrying out her experiments, and being the first to find out something new.
“A lot of my work has involved genetics and making mutants, which entails a lot of PCRs (polymerase chain reactions) and molecular work,” she said. “Day-to-day, you can usually find me in the hood, working with cultures of bacteria and quantifying them on agar plates.”
E. coli might be able to take a break from its Nobel Prize-winning discoveries and make way for V. natriegens, a speedster bacterium with incredible potential for discoveries. Glasgo is particularly excited about its new applications in bioengineering and synthetic biology.